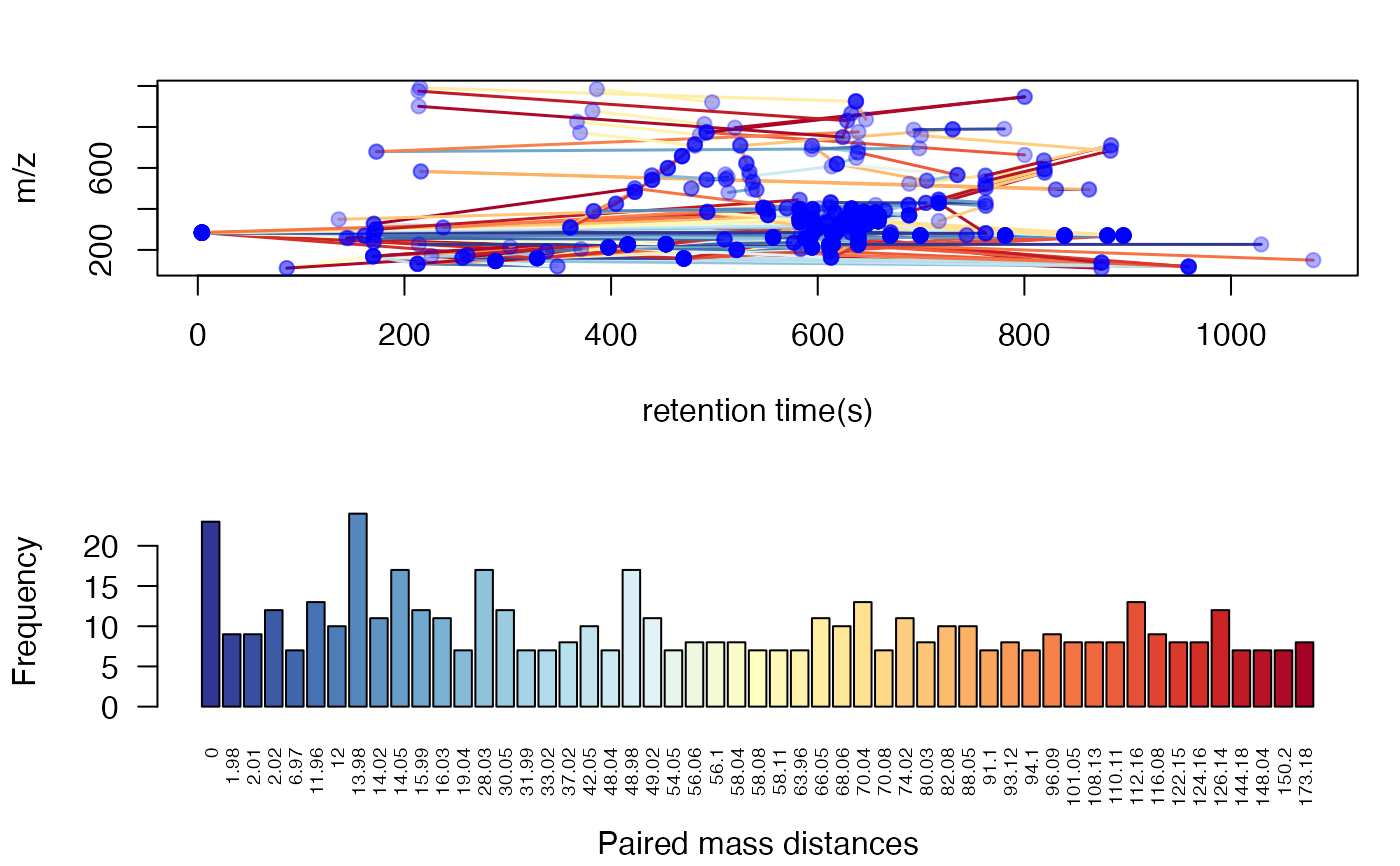
Plot the std mass from GlobalStd algorithm in structure directed analysis(SDA) groups
Source:R/pmdvis.R
plotstdsda.RdPlot the std mass from GlobalStd algorithm in structure directed analysis(SDA) groups
plotstdsda(list, index = NULL, ...)Arguments
See also
Examples
data(spmeinvivo)
re <- globalstd(spmeinvivo, sda=TRUE)
#> 75 retention time clusters found.
#> Using ng = 15
#> 5 unique PMDs retained.
#> The unique within RT clusters high frequency PMD(s) is(are) 28.03 21.98 44.03 17.03 18.01.
#> 409 isotope peaks found.
#> 109 multiple charged isotope peaks found.
#> 251 multiple charged peaks found.
#> 346 paired peaks found.
#> 8 group(s) have single peaks 14 23 32 33 54 55 56 75
#> 11 group(s) with multiple peaks while no isotope/paired relationship 4 5 7 8 11 ... 42 49 68 72 73
#> 9 group(s) with isotope without paired relationship 2 9 22 26 52 62 64 66 70
#> 4 group(s) with paired without isotope relationship 1 10 15 18
#> 43 group(s) with both paired and isotope relationship 3 6 12 13 16 ... 65 67 69 71 74
#> 292 standard masses identified.
#> PMD frequency cutoff is 9 by PMD network analysis with largest network average distance 7.26 .
#> 8 groups were found as high frequency PMD group.
#> 0 was found as high frequency PMD.
#> 2.02 was found as high frequency PMD.
#> 12 was found as high frequency PMD.
#> 28.03 was found as high frequency PMD.
#> 30.05 was found as high frequency PMD.
#> 42.05 was found as high frequency PMD.
#> 58.04 was found as high frequency PMD.
#> 116.08 was found as high frequency PMD.
plotstdsda(re)